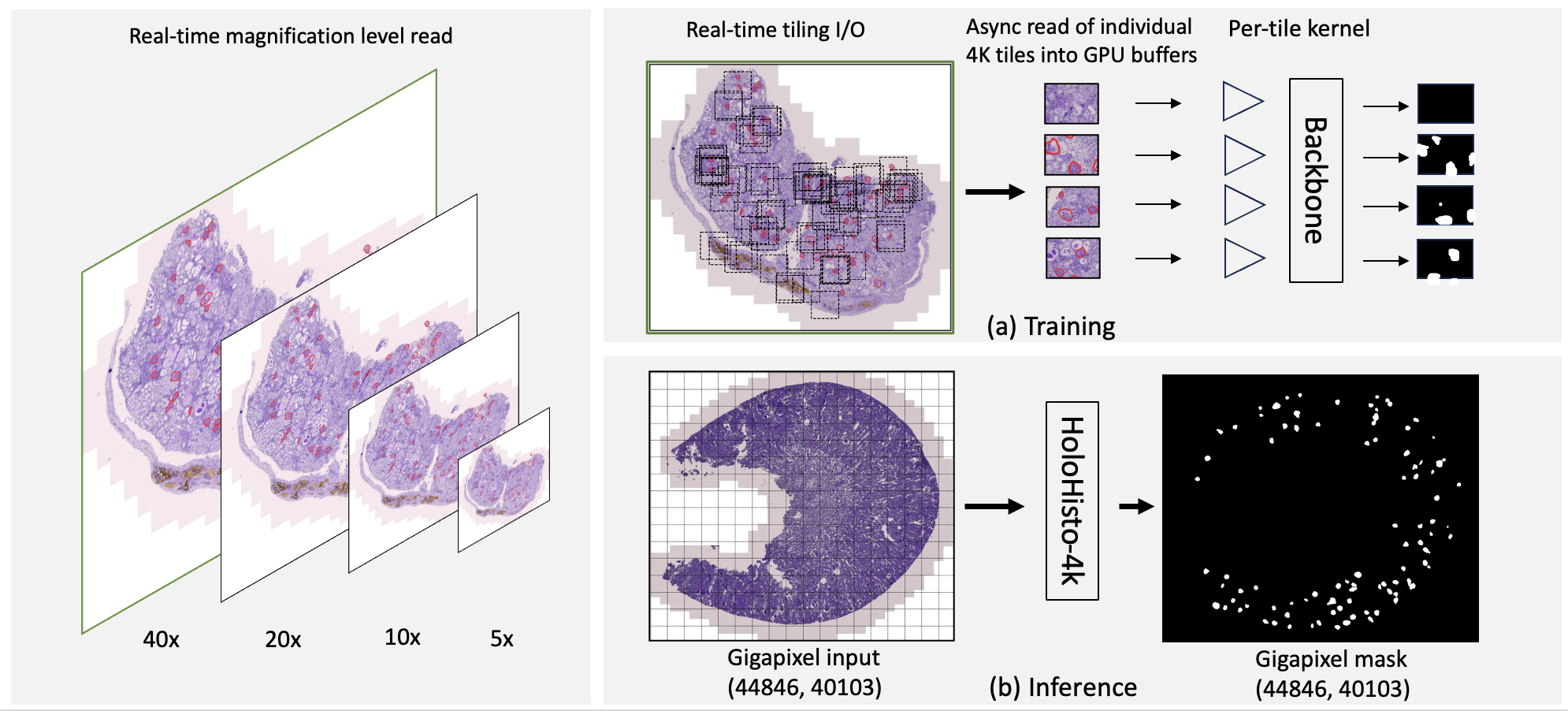
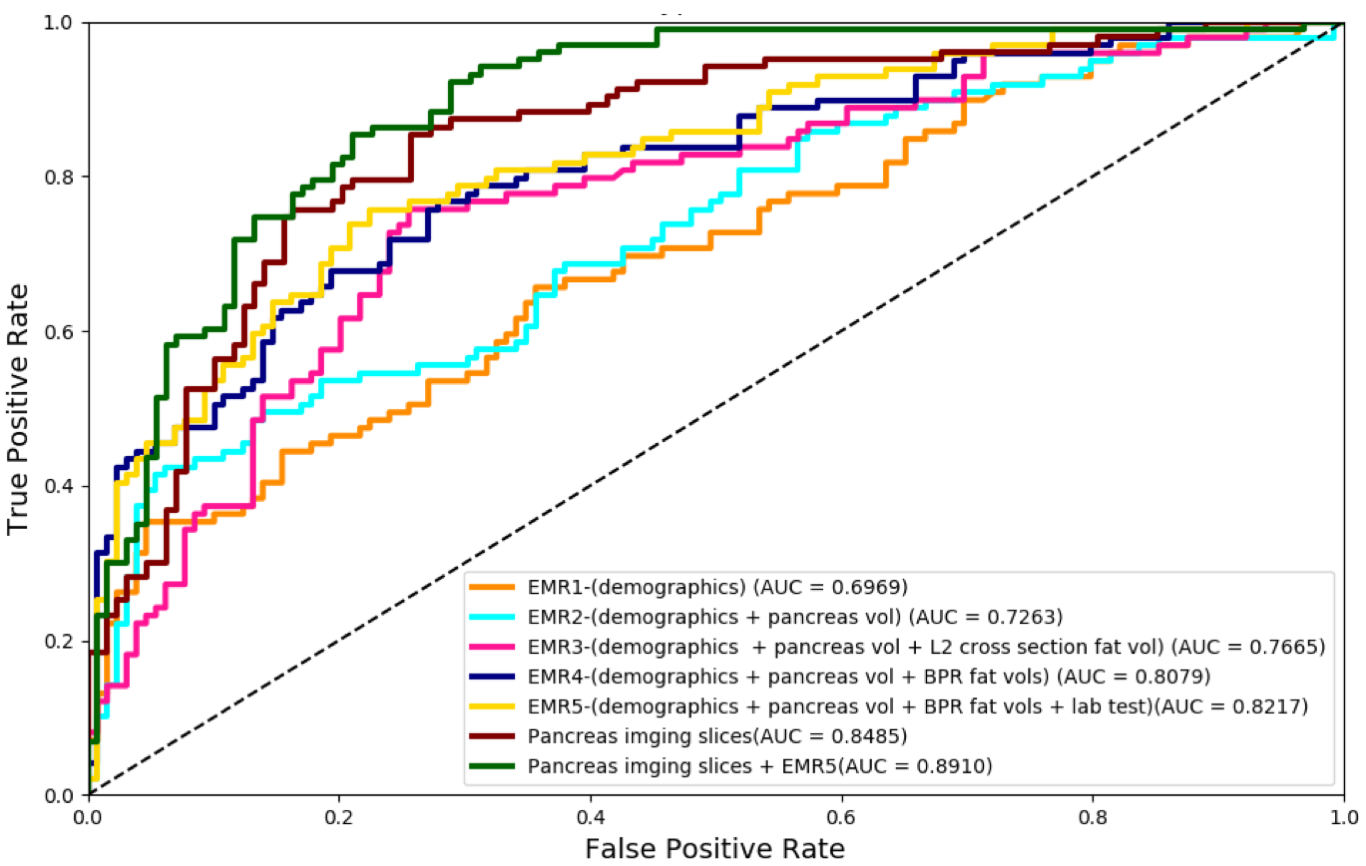
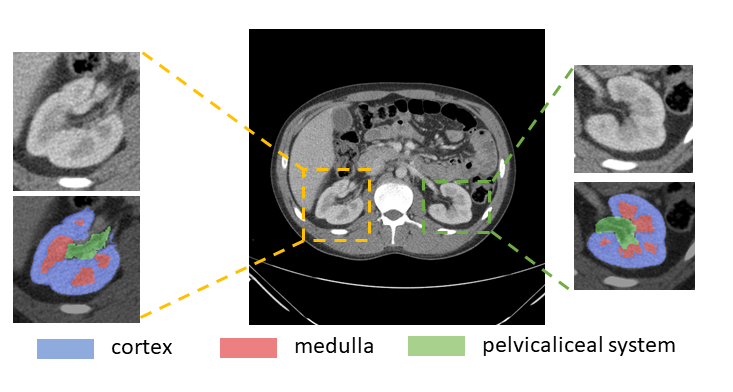
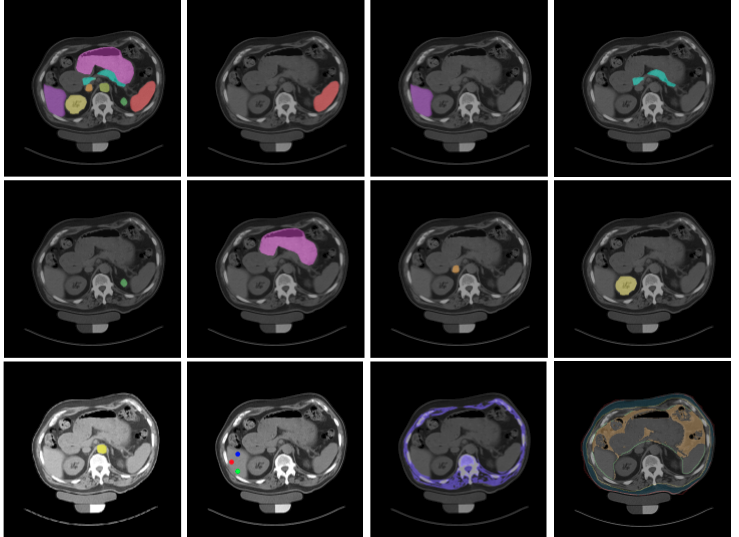
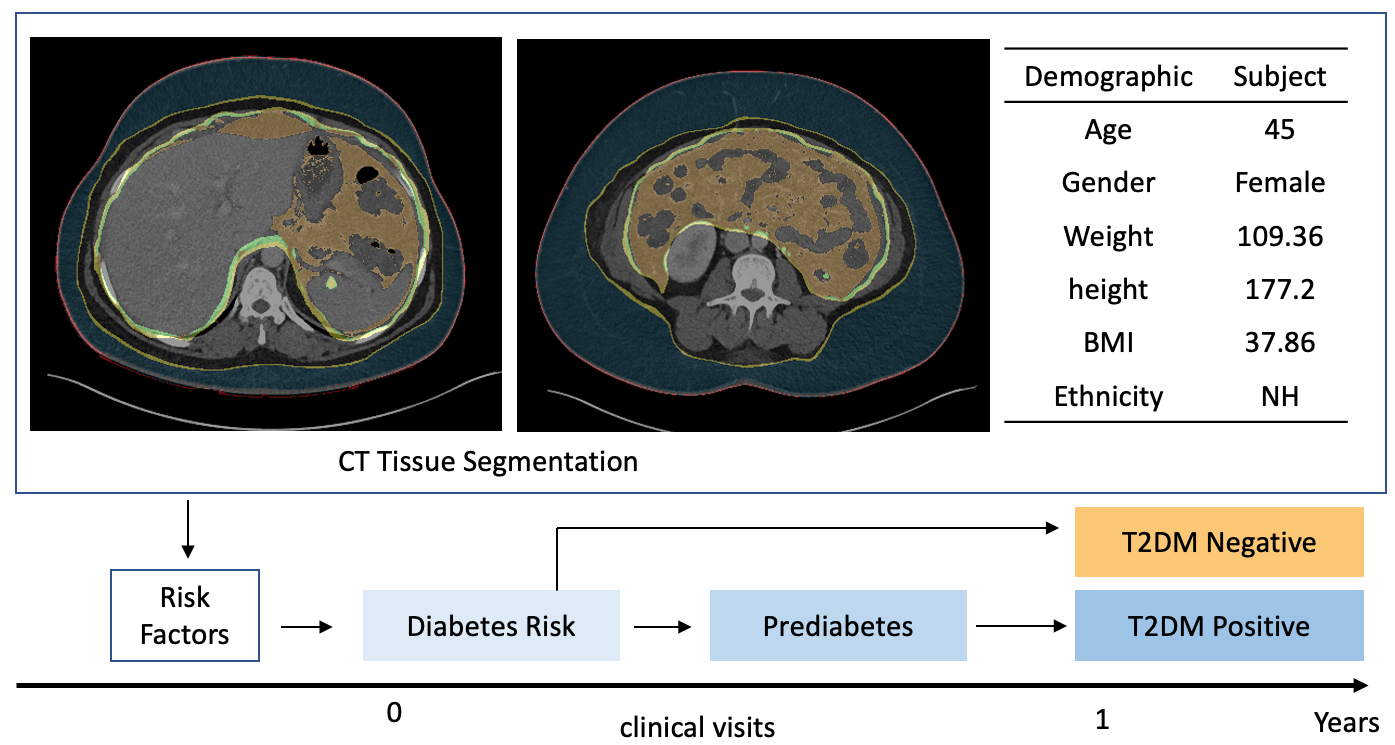
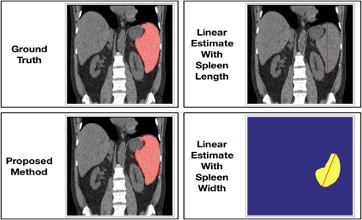
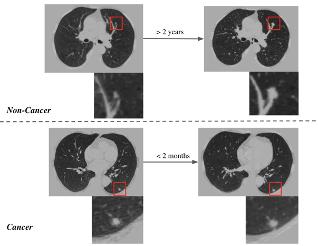
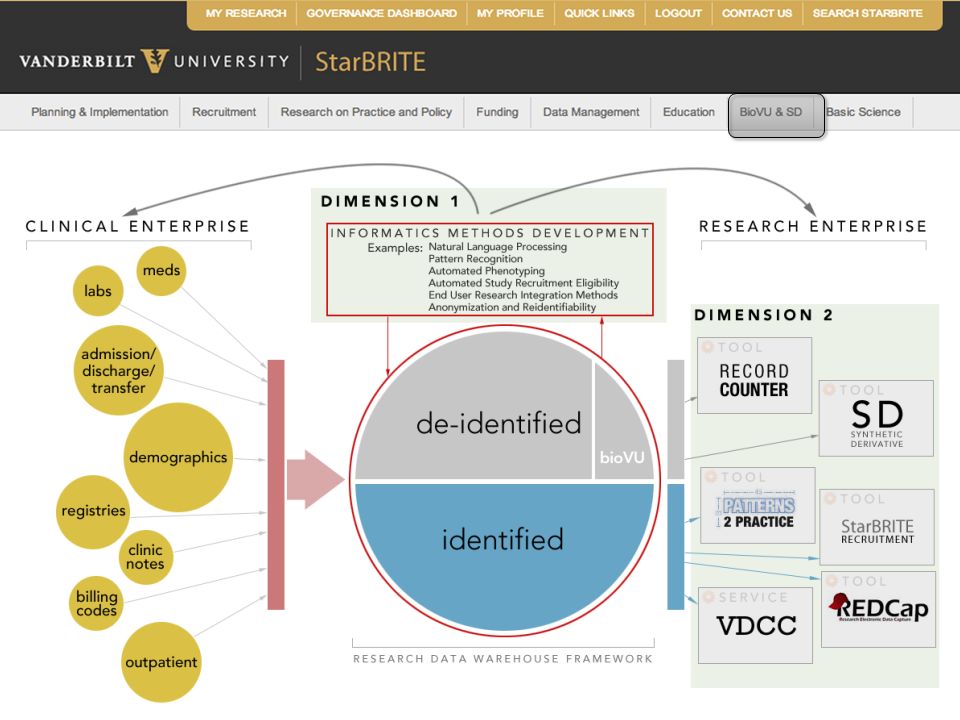
MONAI: Open Healthcare AI Infrastructure
With NVIDIA MONAI cloud APIs, solution providers can more easily integrate AI into their
medical imaging platforms, enabling them to provide supercharged tools for radiologists,
researchers and clinical trial teams to build domain-specialized AI factories. The APIs are
available in Nvidia Cloud Foundation ecosystem through the NVIDIA DGX Cloud AI supercomputing service.